Introduction
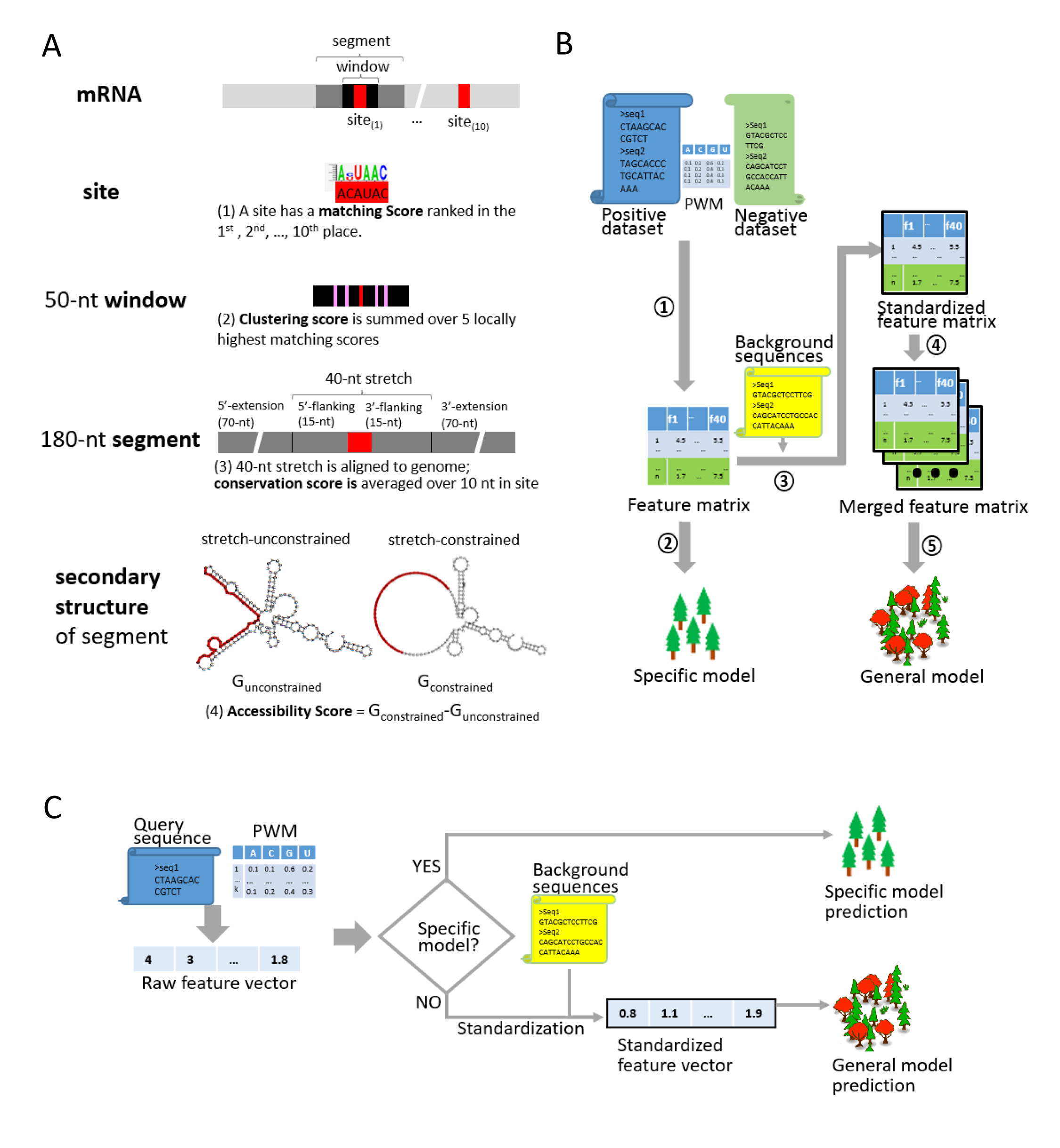
beRBP is a Random Forests based approach to Binding Estimation for human RNA-Binding Proteins (RBP). beRBP is aimed to predict RNA sequence(s) bound by a given RBP characterized with a specific position-weighted matrix (PWM).
beRBP provides a composite solution overarching the RBP-specific and RBP-general strategies. beRBP built 37 ‘Specific models’ for 29 RBPs/37 PWMs, each of which have a decent number of known targets. Meanwhile, it pooled RBP-RNA interactions and trained a ‘General model’ for any RBPs with characterized PWMs. The General model has much broader application scope but comparable performance to Specific models. Currently, beRBP webserver enables binding discovery on one/multiple RNA sequences for 29 RBPs/37 PWMs (Specific models), 143 RBPs/175 PWMs (the General model), and any RBPs with user-provided PWMs or RBP sequences (the General model).
Methodology
Given a candidate RNA sequence and a PWM of a RBP, beRBP generated four types of feature scores, including motif matching, motif clustering, motif conservation, and spatial accessibility. Based on random forests, beRBP-Specific models were trained by known targets of each RBP, while beRBP-General model was built by pooling known targets from all RBPs after post-scoring standardization. beRBP-General captured the common patterns of RBP recognizing targets beyond PWM confinement.