A distribution-free and analytic method for power and sample size calculation in single-cell differential expression
Abstract
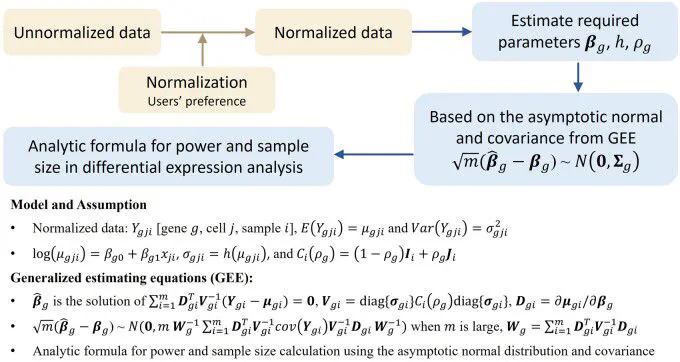
MOTIVATION: Differential expression analysis in single-cell transcriptomics unveils cell type-specific responses to various treatments or biological conditions. To ensure the robustness and reliability of the analysis, it is essential to have a solid experimental design with ample statistical power and sample size. However, existing methods for power and sample size calculation often assume a specific distribution for single-cell transcriptomics data, potentially deviating from the true data distribution. Moreover, they commonly overlook cell-cell correlations within individual samples, posing challenges in accurately representing biological phenomena. Additionally, due to the complexity of deriving an analytic formula, most methods employ time-consuming simulation-based strategies. RESULTS: We propose an analytic-based method named scPS for calculating power and sample sizes based on generalized estimating equations. scPS stands out by making no assumptions about the data distribution and considering cell-cell correlations within individual samples. scPS is a rapid and powerful approach for designing experiments in single-cell differential expression analysis. AVAILABILITY AND IMPLEMENTATION: scPS is freely available at https://github.com/cyhsuTN/scPS and Zenodo https://zenodo.org/records/13375996.