
Manual
HACER has four main functions including browse, query, and download the enhancer data, and contact to report new GRO/PRO/CAGE datasets and to send feedback. This muanual is dedicated to give some guidance to visitors on how to get the most out of the site.
Browse
Users can browse enhancers by cell line. All enhancers in selected cell line(s) are listed in tabular format with the option to filter and customize by annotation.
Detailed information for each enhancer can be revealed by clicking the enhancer ID, which is organized on five tabs for different types of information: basic, TF binding, target genes, GWAS, and eQTLs.
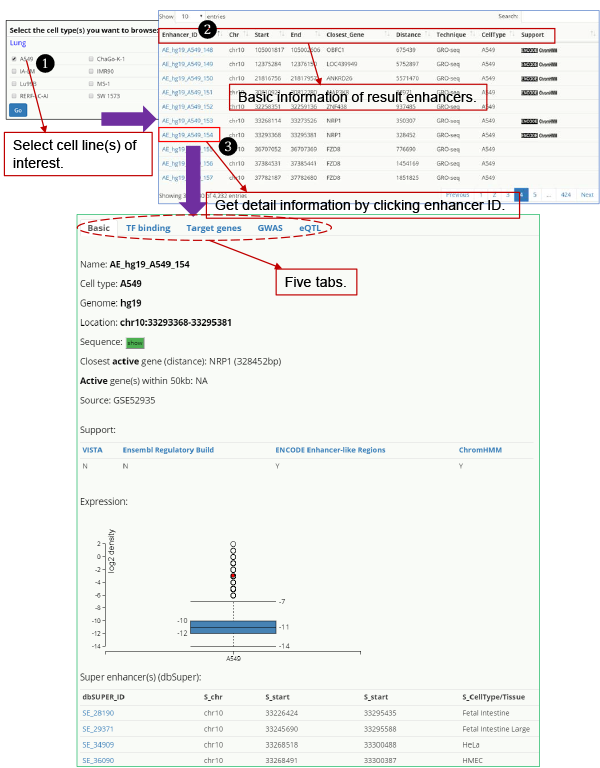
Fig. 1. Browse HACER.
Query
HACER provides four strategies to query the database: SNP-centered, gene-centered, genomic coordinate-centered, and batch query.
SNP-centered When queried on a GWAS risk SNP, HACER returns the cell-type-specific enhancers in which the SNP is located, along with their transcription factor binding and target genes. The transcriptional regulatory network of TF-enhancer-target genes is visualized along with the SNP, providing insight into potential mechanism(s) through which the SNP increases disease risk.
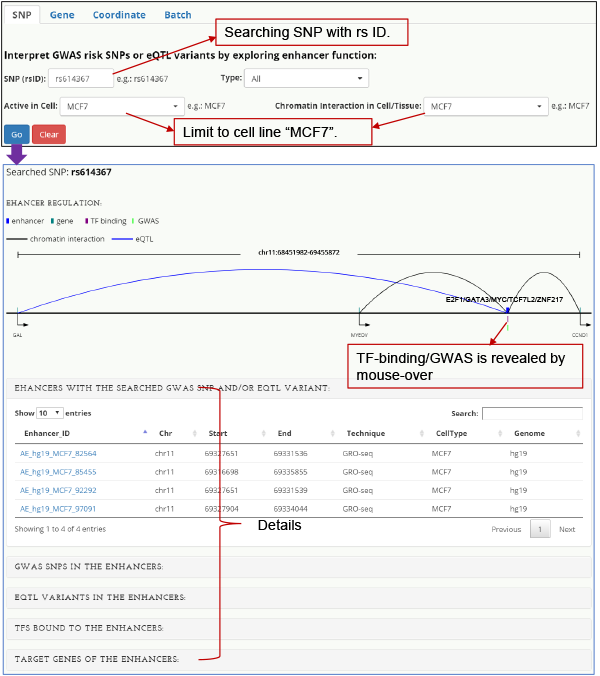
Fig. 2. Interpreting an enhancer risk SNP rs614367.
Gene-centered When queried on a gene, HACER returns all enhancers targeting this gene, providing insight into disease-specific enhancers and enhancer-gene interactions.
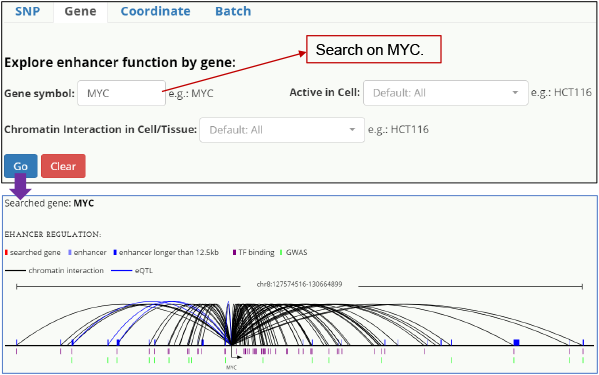
Fig. 3. Enhancer-MYC interaction.
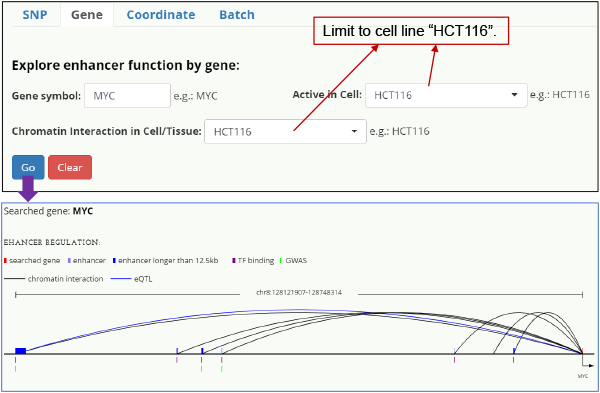
Fig. 4. HCT-116 specific enhancer-MYC interaction. Query on MYC and limited to cell line HCT-116.
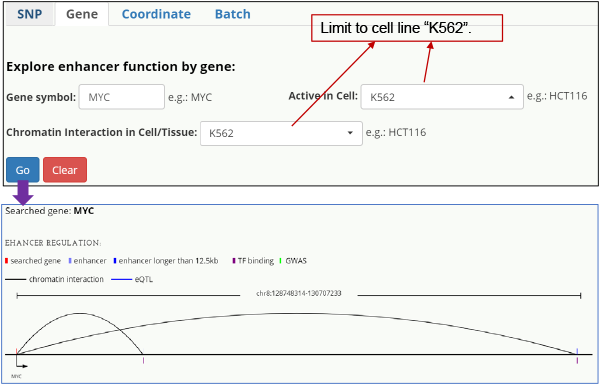
Fig. 5. K562 specific enhancer-MYC interaction. Query on MYC and limited to cell line K562.
Genomic coordinate-centered When queried on a genomic region, HACER finds active enhancers overlapping the region.
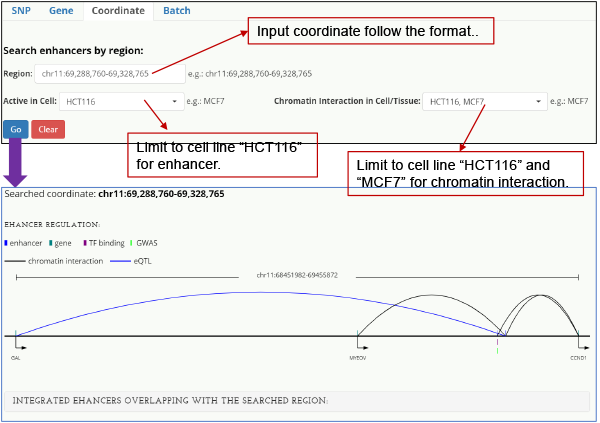
Fig. 6. Genomic coordinate-centered query and result.
Batch query Batch query allows the user to input a set of non-coding variants or enhancers, which HACER then prioritizes by their functional importance.
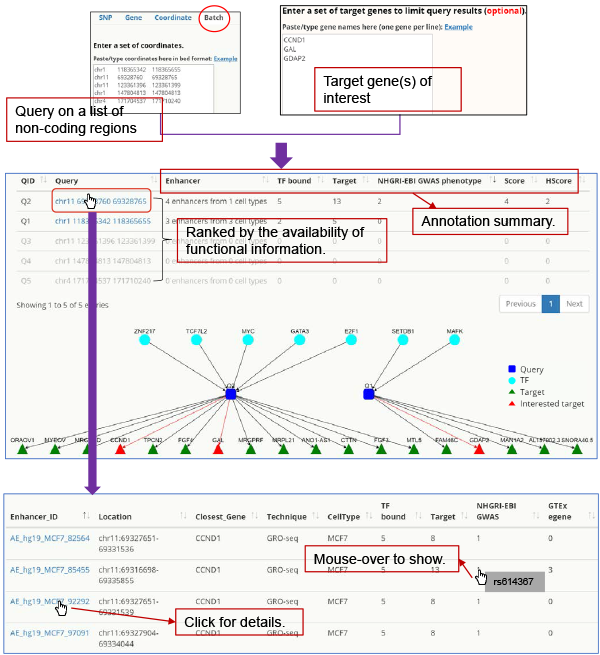
Fig. 7. Prioritizing/annotating non-coding variants/enhancers.
Download
To download enhancers, click on the cell type of interest; the resulting file lists every enhancer in this cell type with comprehensive genomic annotation (e.g., chromatin location, target genes, detection methods).
Contact
HACER provides a Contact page for users to report new PRO-seq, GRO-seq, and CAGE datasets. A hyperlink to the raw data should be provided, from which we will download, and process the data to identify, annotate and deposit enhancers into HACER. New datasets will be processed, and enhancers identified will be updated within HACER. Users also are encouraged to send feedback via the Contact page.